Introduction
Overview
MARTi consists of two main components:
The MARTi Engine - a back-end which performs the analysis and can be a single desktop/laptop or a high performance compute (HPC) cluster.
The MARTi GUI - a lightweight web-based front-end which allows users to view analysis results.
There are a number of possible ways to install MARTi, but in the following sections, we highlight the two most common:
a local analysis configuration where both the MARTi Engine and the MARTi GUI are installed on a single laptop/desktop.
an HPC configuration, where the MARTi Engine resides on an HPC and the MARTi GUI resides somewhere else - e.g. on a laptop/desktop.
Local analysis configuration
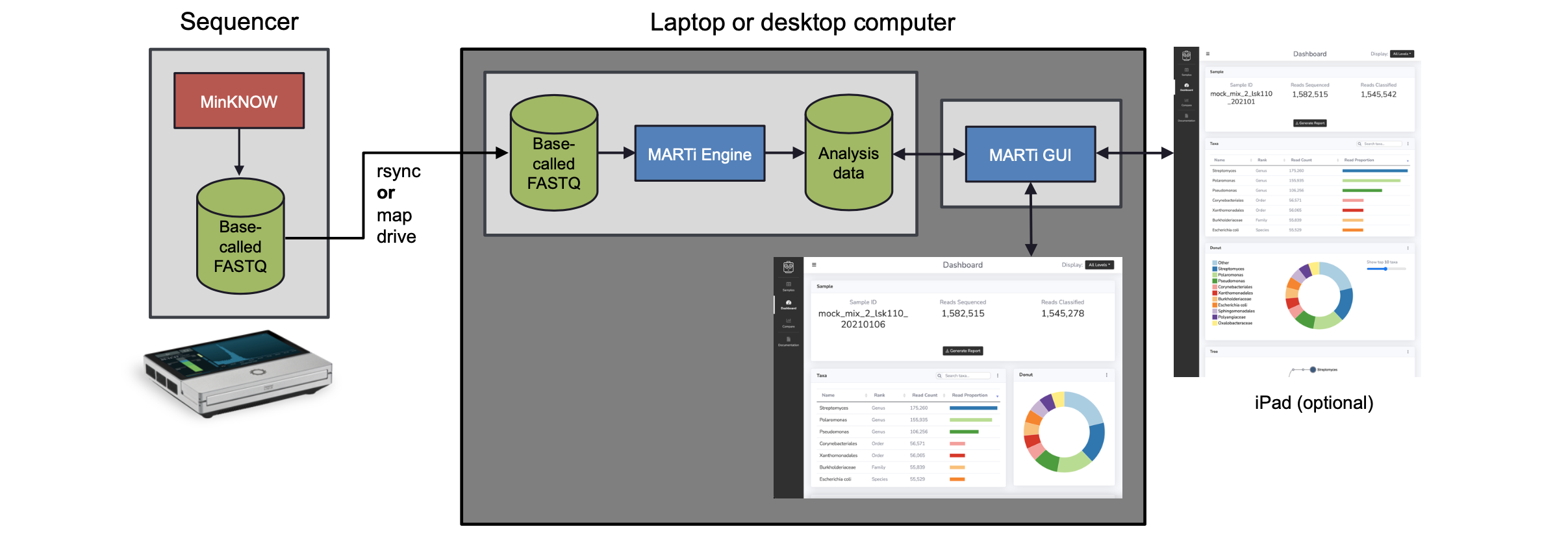
In the local analysis configuration:
A sequencing device (e.g. MinION Mk1C or GridION) generates basecalled reads.
On a laptop or desktop computer, the MARTi Engine analyses the sequence data. It accesses the data either by mapping drives or rsync’ing the data over a network connection.
On the same computer, the MARTi GUI provides a server which enables web browsers to access analysis. This could be a browser on the same computer, or any browser on the same network.
HPC configuration
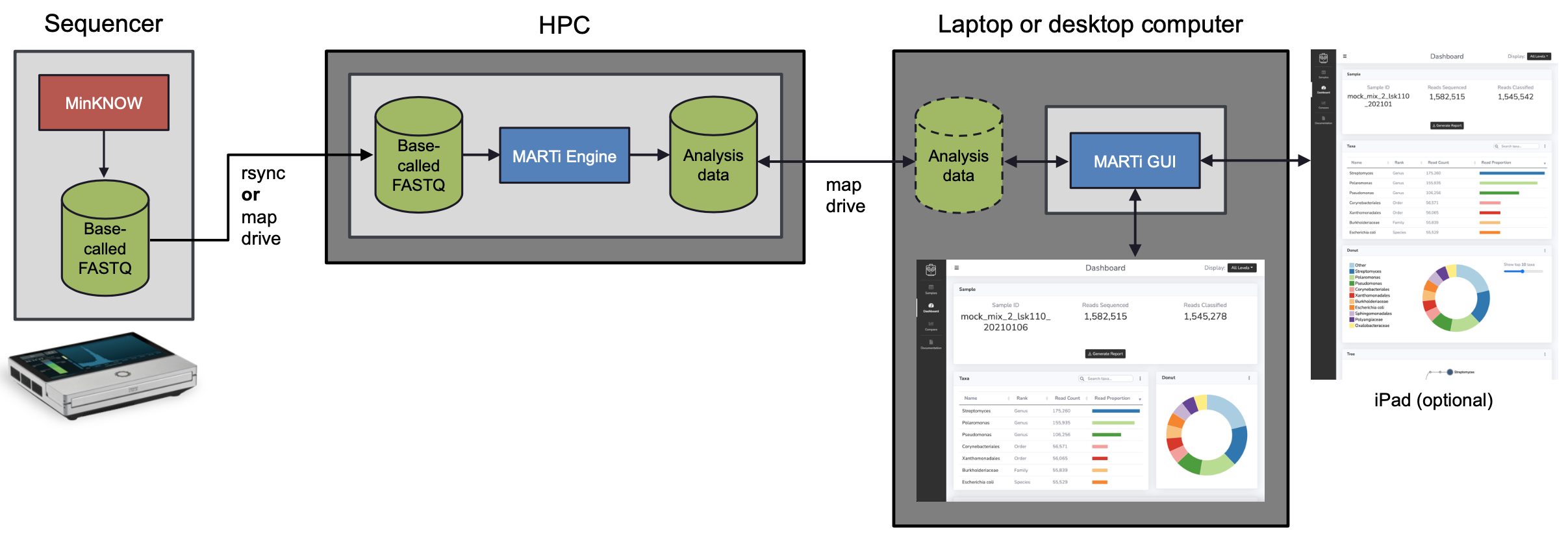
In the HPC configuration:
A sequencing device (e.g. MinION Mk1C or GridION) generates basecalled reads.
On the HPC system, the MARTi Engine analyses the sequence data. It accesses the data either by mapping drives or rsync’ing the data over a network connection.
On a laptop or desktop, the MARTi GUI provides a server which enables web browsers to access analysis. This could be a browser on the same computer, or any browser on the same network.
Inside MARTi Engine
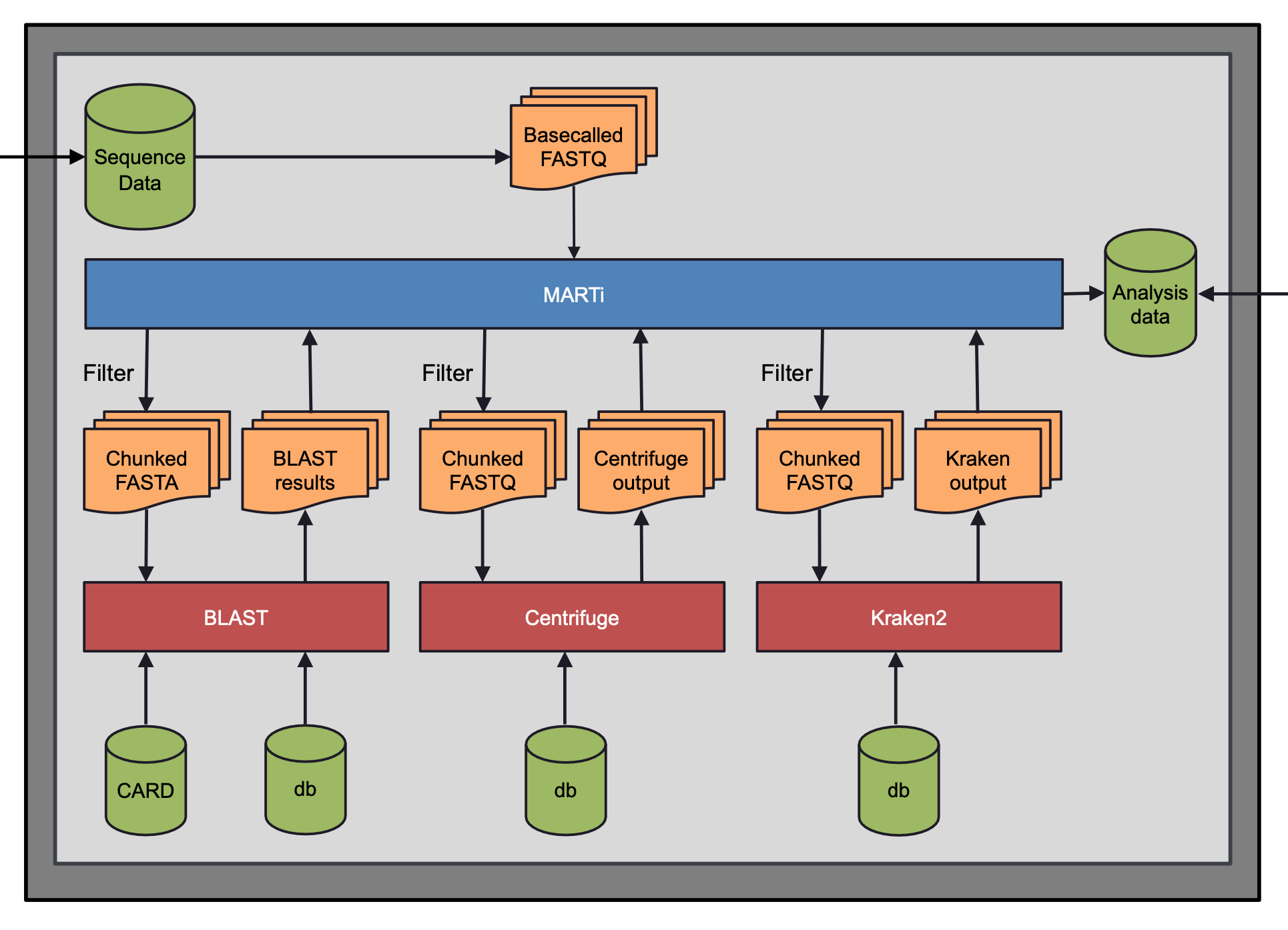
The diagram above illustrates the processes that take place inside the MARTi Engine:
Basecalled reads first pass through a prefilter which removes reads that don’t meet a quality or length threshold.
Remaining reads are batched into chunks for speedy analysis.
By default, MARTi classifies reads with a combination of BLAST and its own Lowest Common Ancestor algorithm to assign reads to taxa. Centrifuge and Kraken2 can also be used as an alternative for lower powered systems.
BLAST files are gzip compressed after analysis to reduce storage requirements.
For AMR classification, MARTi uses BLAST and the CARD database.